Large-scale chemical cross-linking with mass spectrometry (XL-MS) analyses are quickly becoming a powerful means for high-throughput determination of protein structural information and protein-protein interactions. Recent studies have garnered thousands of cross-linked interactions, yet the field lacks an effective tool to compile experimental data or access the network and structural knowledge for these large scale analyses.
We present XLinkDB which integrates tools for network analysis, Protein Databank queries, modeling of predicted protein structures and modeling of docked protein structures. By incorporating cross-linking data, and model scoring scripts (e.g. XLMap), this platform provides the means for experts and those new to XL-MS to identify interesting interactions and analyze the structural landscape predicted by XL-MS data. The novel, integrated approach of XLinkDB enables the holistic analysis of XL-MS protein interaction data without limitation to the cross-linker or analytical system used for the analysis.
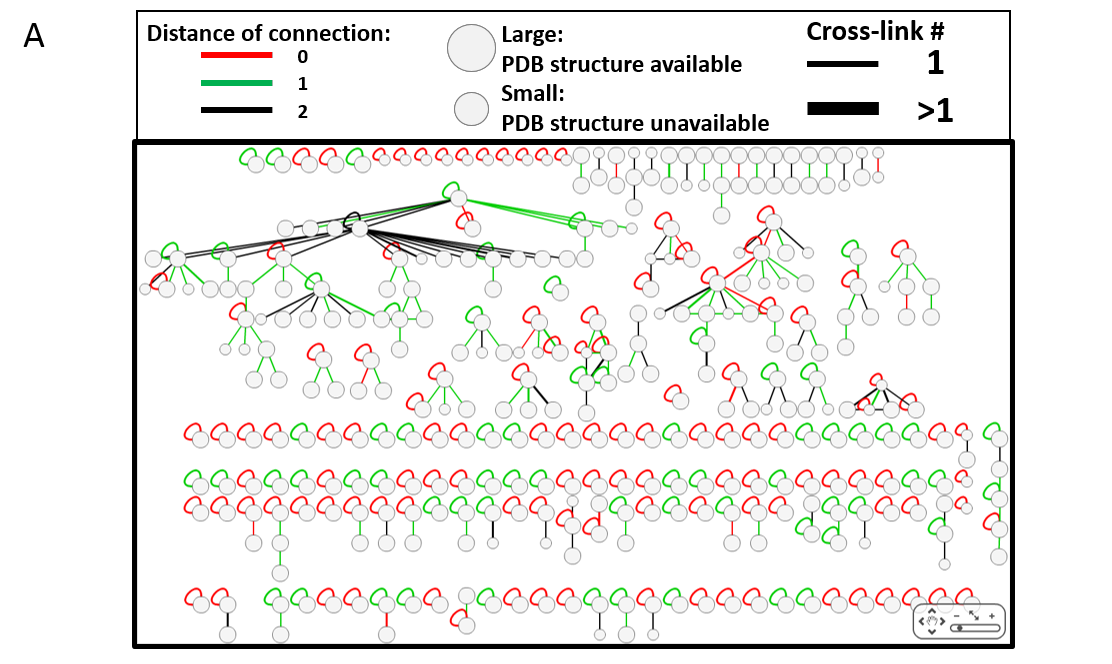
Each XL-MS network on XLinkDB comes from published works using a variety of different cross-linker chemistries across multiple model organisms (see Citations). Nodes within these networks represent individual proteins identified by XL-MS. They may have been identified as cross-linked only to themselves (self loops) or in interactions with other proteins. The edges connecting each node represent one or more cross-link identifications. Often, these cross-links have site level information that can be accessed using the Table View function for a given netowrk.
Networks allow for global analysis of the interactomes for each dataset or species. XlinkDB extends this network analysis by automating the next phase of XL-MS analysis, Structural Analysis.
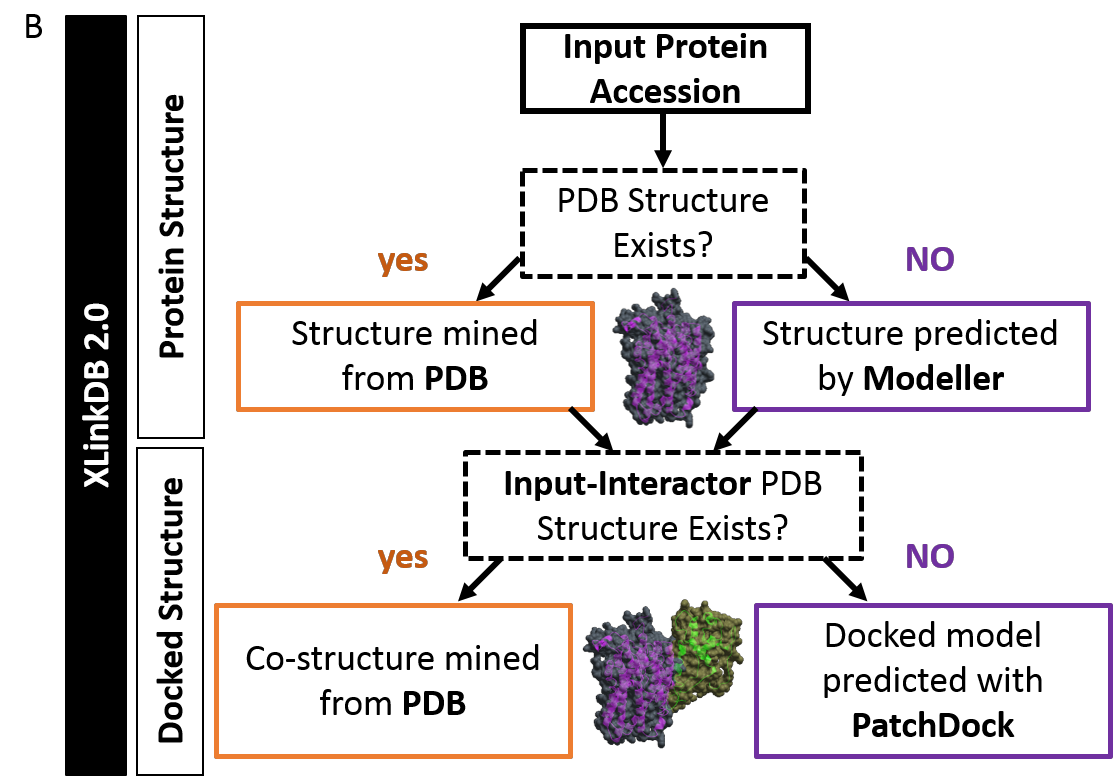
The Protein Databank contains thousands of protein structures that can aid in functional analysis of proteins which we are able to mine and integrate with XL-MS data within XlinkDB . Yet, many proteins identified by proteomic experiments have limited or no structural information. In an effort to improve the structural insights that researchers can attain from XL-MS data, we developed a pipeline that automates the mining, modeling, and docking of cross-linked protein interactions.
Modeling and docking operations within XlinkDB are performed using the Integrative Modeling Platform from the Sali Lab. Modeling is performed using MODELLER and docking is performed using PatchDock based on cross-linked distance constraints.
With the increasing number of XL-MS studies rich in network and structural information, XLinkDB now allows for organism-wide network creation. In this way it is possible to generate protein interactions networks from the sum of multiple curated Xl-MS studies (e.g. a full human XL-MS network). These summed, species-level networks can enable proteome-scale network analysis of protein interactions. Moreover, because the data is derived from XL-MS studies each of these network nodes still contain a wealth of structural information for thousands of proteins across multiple organisms.